Graph Sampling¶
Graph sampling is an effective technique for managing large graphs and is widely used in programming paradigms represented by the GraphSAGE framework. Sampling reduces data size and facilitates efficient processing by Tensor-Based computing frameworks through data alignment.
Before sampling, we are required to provide seeds, which can be either nodes or edges. Correspondingly, GLE provides graph traversal operators to prepare the seeds for batch-sampling:
node sampler
edge sampler
Then, sampling requirements by users can be abstracted and categorized into the following classes of operations in GLE:
Neighborhood Sampling.
Subgraph Sampling.
Negative Sampling.
Neighborhood Sampling involves selecting one-hop or multi-hop neighboring vertices based on the input vertex to construct the perceptual field in GCN theory. Input for neighborhood sampling can be from the graph traversal output or other external data sources.
Subgraph Sampling involves vertices of one-hop or multi-hop and all edges with src and dst vertices already sampled, which form a subgraph. This sampling method becomes more and more important as the research in GNN expressivity advances.
Negative Sampling selects vertices that are not directly connected to the input vertices, and it is commonly used in unsupervised learning.
Graph Traversal¶
Graph traversal, in GNN, has a different semantics than classical graph computation. The training model of mainstream deep learning algorithms iterates by batch. To meet this requirement, the data has to be accessible by batch, and we call this data access pattern traversal. In GNN algorithms, the data source is the graph, and the training samples usually consist of the vertices and edges of the graph. Graph traversal refers to providing the algorithm with the ability to access vertices, edges or subgraphs by batch.
Currently GLE supports batch traversal of vertices and edges. This random traversal can be either putback-free or putback. In a no-replay traversal, gl.OutOfRangeError is triggered every time an epoch ends. The data source being traversed is partitioned, i.e. the current worker (in the case of distributed TF) only traverses the data on the Server corresponding to it.
For usage and interfaces of graph traversal, please check the traversal part of GLE for more details.
Neighborhood Sampling¶
Different implementation strategies such as random and edge-weight are available for each sampling operation. We have accumulated over 10 sampling operators through practical production and have made the operator programming interface open to allow user customization to keep up with the ever-evolving GNN.
For sampling strategies, GLE currently has support for the following sampling strategies, corresponding to the strategy parameters when generating NeighborSampler objects.
strategy |
description |
|---|---|
edge_weight |
Samples with probability with edge weights |
random |
random_with_replacement |
topk |
Return the neighbors with edge weight topK, if there are not enough neighbors, refer to the padding rule. |
in_degree |
Probability sampling by vertex degree. |
full |
Returns all neighbors, the expand_factor parameter does not work, the result object is SparseNodes or SparseEdges. |
Next is an example for neighbor sampling:
Example:
As shown in the figure below, starting from a vertex of type user, sample its 2-hop neighbors, and return the result as layers, which contains layer1 and layer2. layer’s index starts from 1, i.e. 1-hop neighbor is layer1 and 2-hop neighbor is layer2.
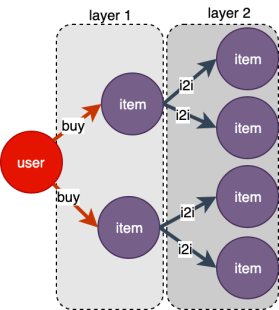
s = g.neighbor_sampler(["buy", "i2i"], expand_factor=[2, 2])
l = s.get(ids) # input ids: shape=(batch_size)
# Nodes object
# shape=(batch_size, expand_factor[0])
l.layer_nodes(1).ids
l.layer_nodes(1).int_attrs
# Edges object
# shape=(batch_size * expand_factor[0], expand_factor[1])
l.layer_edges(2).weights
l.layer_edges(2).float_attrs
For usage and interfaces of neighborhood sampling, please check the sampling part of GLE for more details.
Subgraph Sampling¶
Unlike EgoGraph, SubGraph contains the edge_index of the graph topology, so the message passing path (forward computation path) can be determined directly by the edge_index, and the implementation of the conv layer can be done directly by the edge_index and the nodes/edges data. In addition, SubGraph is fully compatible with the Data in PyG, so the model part of PyG can be reused.
Subgraph Sampling involves vertices of one-hop or multi-hop and all edges with src and dst vertices already sampled, which form a subgraph.
This sampling method becomes more and more important as the research in GNN expressivity advances.
Negative Sampling¶
Negative sampling refers to sampling vertices that have no direct edge relationship with a given vertex. Similar to neighbor sampling, negative sampling has different implementation strategies, such as random, in-degree of nodes, etc. As a common operator of GNN, negative sampling supports extensions and scenario-oriented customization. In addition, GLE provides the ability to negative sampling by specified attribute condition.
GLE currently supports the following negative sampling strategies, corresponding to the strategy argument when generating NegativeSampler objects.
strategy |
description |
|---|---|
random |
Random negative sampling, not guaranteed true-negative |
in_degree |
Negative sampling with probability of vertex entry distribution, guaranteed true-negative |
node_weight |
Negative sampling with probability of vertex weight, true-negative |
Next is an example for negative sampling:
Example:
es = g.edge_sampler("buy", batch_size=3, strategy="random")
ns = g.negative_sampler("buy", 5, strategy="random")
for i in range(5):
edges = es.get()
neg_nodes = ns.get(edges.src_ids)
print(neg_nodes.ids) # shape is (3, 5)
print(neg_nodes.int_attrs) # shape is (3, 5, count(int_attrs))
print(neg_nodes.float_attrs) # shape as (3, 5, count(float_attrs))
For usage and interfaces of negative sampling, please check the negative sampling part of GLE for more details.
GSL¶
GLE abstracts sampling operations into a set of interfaces, called GSL (Graph Sampling Language). Generally, graph sampling consists of several categories as follows.
Traversal type (Traverse), which obtains point or edge data of a batch from the graph.
Relational (Neighborhood, Subgraph), which obtains the N-hop neighborhood of points or generates a subgraph composed of points for constructing training samples.
Negative sampling (Negative), as opposed to relational, which is generally used in unsupervised training scenarios to generate negative example samples.
For example, for the heterogeneous graph scenario of “users clicking on products”, “randomly sample 64 users and sample 10 related products for each user by the weight of the edges”. This can be presented by GSL as
g.V("user").batch(64).outV("click").sample(10).by("edge_weight")
GSL covers support for oversized graphs, heterogeneous graphs, and attribute graphs considering the characteristics of the actual graph data, and the syntax is designed close to the Gremlin form for easy understanding.
For more details, please check the GSL part of GLE.